Chapter 20 Fitting curves cribsheet
20.1 Fitting polynomials
20.2 The data
d<-read.csv("/home/aqm/course/data/marineinverts.csv")
DT::datatable(d)20.3 Fitting linear model
20.3.1 Linear model fit
mod<-lm(data=d,richness~grain)20.3.2 Linear model anova and summary
anova(mod)## Analysis of Variance Table
##
## Response: richness
## Df Sum Sq Mean Sq F value Pr(>F)
## grain 1 385.13 385.13 23.113 1.896e-05 ***
## Residuals 43 716.52 16.66
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod)##
## Call:
## lm(formula = richness ~ grain, data = d)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6.8386 -2.0383 -0.3526 2.5768 11.6620
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 18.669264 2.767726 6.745 3.01e-08 ***
## grain -0.046285 0.009628 -4.808 1.90e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.082 on 43 degrees of freedom
## Multiple R-squared: 0.3496, Adjusted R-squared: 0.3345
## F-statistic: 23.11 on 1 and 43 DF, p-value: 1.896e-0520.3.3 Linear model plot
library(ggplot2)
theme_set(theme_bw())
g0<-ggplot(d,aes(x=grain,y=richness))
g1<-g0+geom_point() + geom_smooth(method="lm")
g1 + xlab("Mean grain size") + ylab("Species richness")
20.3.4 Reset test
We can check whether a strait line is a good representation of the pattern using the reset test that will have a low p-value if the linear form of the model is not a good fit.
library(lmtest)
resettest(d$richness ~ d$grain)##
## RESET test
##
## data: d$richness ~ d$grain
## RESET = 19.074, df1 = 2, df2 = 41, p-value = 1.393e-0620.3.5 Durbin Watson test
The Durbin Watson test which helps to confirm serial autocorrelation that may be the result of a misformed model will often also be significant when residuals cluster on one side of the line. In this case it was not, but this may be because there were too few data points.
dwtest(d$richness~d$grain)##
## Durbin-Watson test
##
## data: d$richness ~ d$grain
## DW = 1.7809, p-value = 0.1902
## alternative hypothesis: true autocorrelation is greater than 020.3.6 Diagnostic plot after fitting linear model
library(ggfortify)## Error in library(ggfortify): there is no package called 'ggfortify'theme_set(theme_bw())
autoplot(mod, which = 1)## Error: Objects of type lm not supported by autoplot.20.4 Fitting quadratic model
20.4.1 Quadratic model fit
mod2<-lm(data=d,richness~grain + I(grain^2))20.4.2 Quadratic model anova and summary
anova(mod2)## Analysis of Variance Table
##
## Response: richness
## Df Sum Sq Mean Sq F value Pr(>F)
## grain 1 385.13 385.13 29.811 2.365e-06 ***
## I(grain^2) 1 173.93 173.93 13.463 0.00068 ***
## Residuals 42 542.59 12.92
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod2)##
## Call:
## lm(formula = richness ~ grain + I(grain^2), data = d)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6.5779 -2.5315 0.2172 2.1013 8.3415
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 53.0133538 9.6721492 5.481 2.21e-06 ***
## grain -0.2921821 0.0675505 -4.325 9.19e-05 ***
## I(grain^2) 0.0004189 0.0001142 3.669 0.00068 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.594 on 42 degrees of freedom
## Multiple R-squared: 0.5075, Adjusted R-squared: 0.484
## F-statistic: 21.64 on 2 and 42 DF, p-value: 3.476e-0720.4.3 Quadratic model plot
library(ggplot2)
theme_set(theme_bw())
g0<-ggplot(d,aes(x=grain,y=richness))
g1<-g0+geom_point() + geom_smooth(method="lm", formula=y~x+I(x^2), se=TRUE)
g1 + xlab("Mean grain size") + ylab("Species richness")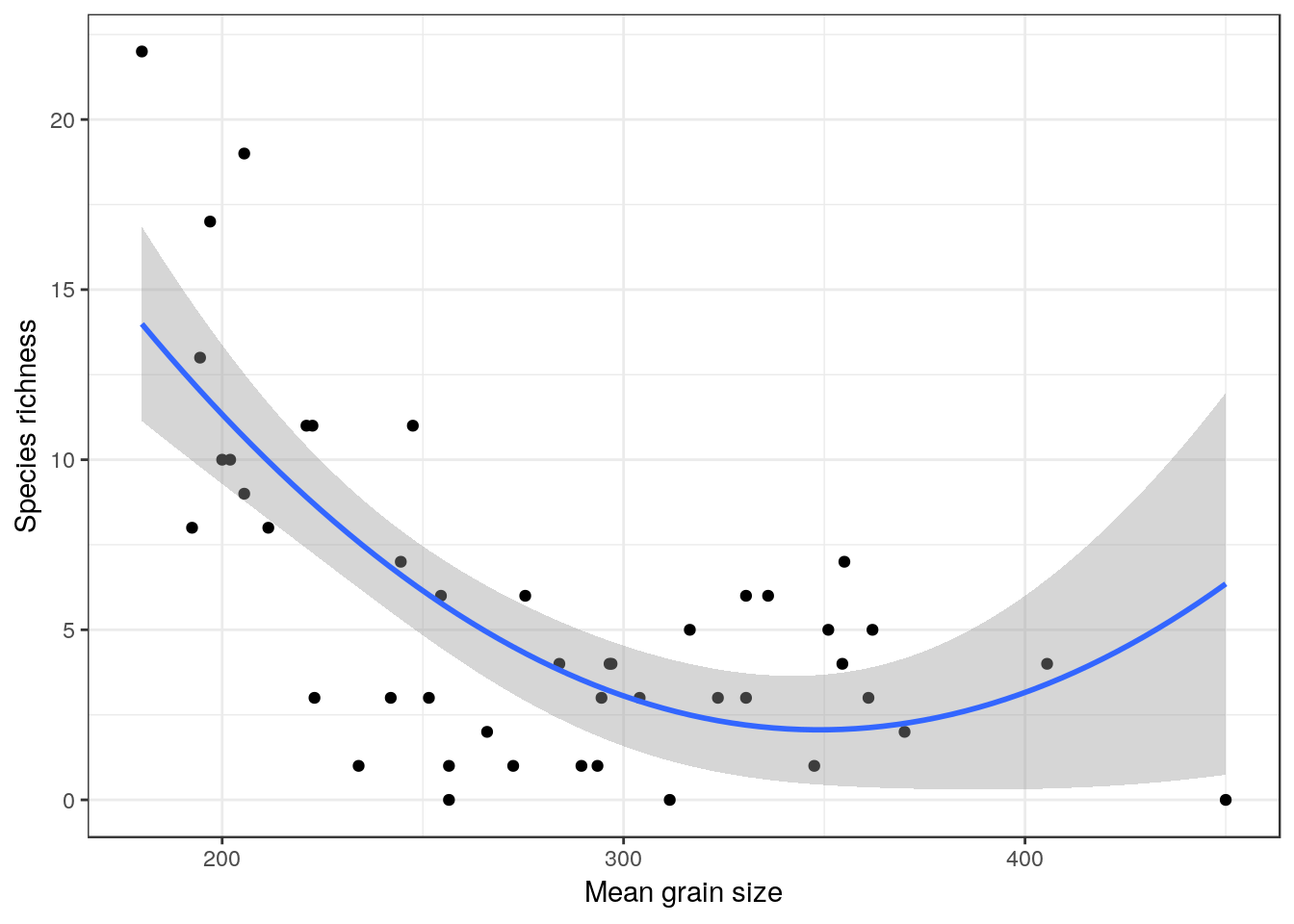
20.4.4 Diagnostic plot after fitting quadratic
library(ggfortify)## Error in library(ggfortify): there is no package called 'ggfortify'theme_set(theme_bw())
autoplot(mod2, which = 1)## Error: Objects of type lm not supported by autoplot.20.4.5 Comparing fits
anova(mod,mod2)## Analysis of Variance Table
##
## Model 1: richness ~ grain
## Model 2: richness ~ grain + I(grain^2)
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 43 716.52
## 2 42 542.59 1 173.93 13.463 0.00068 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1